 HIGH RESOLUTION spectral precision distance microscopy reveals
three-dimensional positions of active and inactive ANT-genes on the female
X-chromosome.
HIGH RESOLUTION spectral precision distance microscopy reveals
three-dimensional positions of active and inactive ANT-genes on the female
X-chromosome. BIOPHYSICS_GENOME NANOSCOPY
Geometry Matters
Optical microscopy reveals three-dimensional genome-nanostructures
By Georg Wolschin
The Humane Genome Project's main goal is to determine the sequence of the four letters of the genetic alphabet, and it has recently suceeded to do so on chromosomes 21 and 22, with a total of at least 770 genes. Alas, the linear sequence alone does not provide all the information a cell needs in order to function properly. In particular, the complicated three-dimensional geometrical structure of the DNA-molecule within each of the 46 chromosomes is expected to be very important for the functioning of the cell.
Which of the genes are active could, for example, significantly be influenced by geometry. Hence, the time-dependent position of a gene close to, or farther inside, the three-dimensional extend of the chromosome is very relevant. However, the resolution of conventional optical microscopes is not good enough to reveal such small spatial structures on a nanometer scale in cell nuclei.
Generally speaking, the spatial resolution of optical microscopes ceases at the so-called Abbe-limit, which is proportional to the wavelength of the light, and the aperture angle. At maximum aperture of about 70 degrees, the microscope's focus is about 200 nanometers wide and 700 nanometers long. This is much more than typical distances between functionally relevant genome-microstructures - for example, active genes, and the surface of chromosomes.
Although high-resolution schemes such as x-ray and neutron crystallography have provided important structural informations on many biological macromolecules, they are not well-suited for the genome, because they may split off genes and break chromosomes due to their higher energy. In addition, they have to rely on probes in vacuum and are thus not usable for living cells. Newly developed methods of optical microscopy which allow to investigate nanostructures of intact cell nuclei are therefore of utmost interest.
As a surprise, the resolution of optical microscopes has been drastically improved far beyond the Abbe-limit through interference of two opposed wave-fronts at the Max-Planck Institute for biophysical research in Germany. Stefan Hell and his colleagues attain almost the same resolution as in case of an ideal microscope, which would have an aperture of 360 degrees. The new setup is thus called 4Pi-confocal microscope. It has the best resolution of all currently available optical microscopes, and it can contribute to reveal nanostructures in cell nuclei.
Moreover, a completely new method to improve optical resolution has been developed that is based on optical markers: Small DNA sequences are labelled in intact cell nuclei by fluorescence. With a high-resolution light microscope and digital image processing, the center of a labelled site can then be localized with a precision which is much better than the optical resolution of the microscope that has been used. A Laser-scanning microscope, for example, thus yields the three-dimensional position of a gene with a precision of 10 nanometers - which is about 1/50 of the wavelength of the incident light, and far beyond Abbe's limit. With the 4Pi-microscope, which is still in the experimental stage, it would be even better.
Now, If two closely neighboring sites - for example, two genes - carry markers that differ in their spectral signatures, their individual positions in space can be registered independently of each other. As a consequence, their distance in three dimensions can be determined, even though it may be much smaller than the optical resolution of the microscope that is being used. The method is called Spectral Precision Distance Microscopy (SPDM) and has been developed by Christoph Cremer and his group at Heidelberg University. It allows to resolve objects of different spectral signature in arbitrary positions in three-dimensional space down to a distance of presently about 30 nanometers, with a considerable potential for further improvement. It is thus optimally suited to investigat e complex 3d-nanostructures of the chromosomes far beyond the usual optical resolution limit.
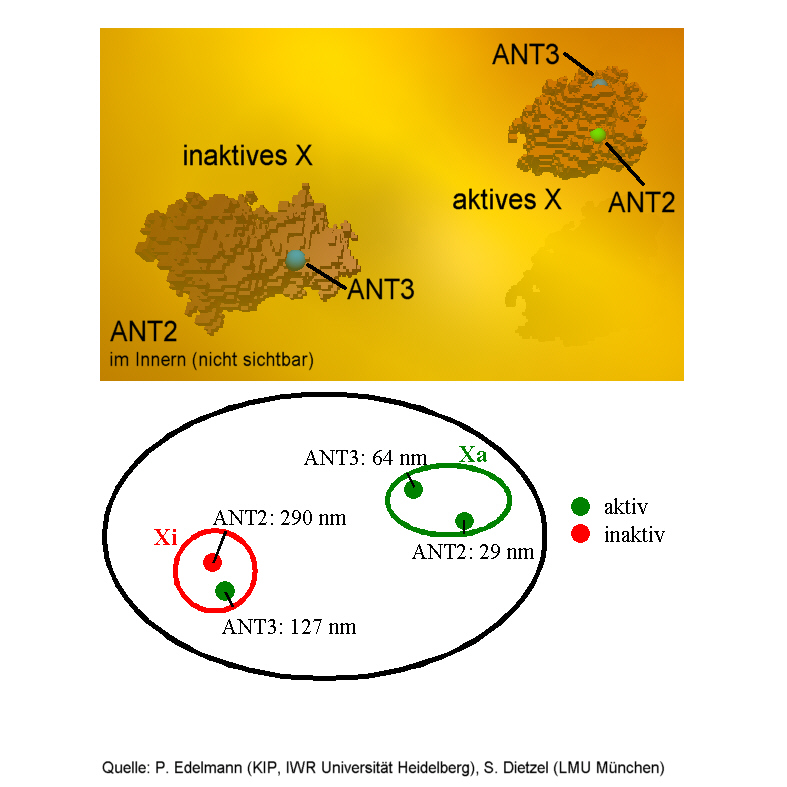
First exploratory measurements have been carried out to study the positions of two specific genes - the so-called Adenine Nucleotide Transferase genes ANT2 and ANT3 - which are localized on the female X-chromosome. The experimentalists could verify the hypothesis that, at least in this case, gene activation corresponds to a change in the three-dimensional structure of the relevant part of the chromosome. As compared to the active gene, the inactive one has moved by typically 260 nanometers into the interior of the chromosome and is, therefore, not attainable for certain proteins. Hence, time-dependent geometrical changes are likely to supplement the role of known elements such as activator proteins in the mechanisms of gene regulation: a clear proof of the importance of spatial genome nanostructure.
In view of such results, efforts are underway to improve on the smallest distance that can be resolved with SPDM. This requires to further enhance the normal optical resolution as indicated by the 4Pi-microscope on the one hand, and to improve the labelling and image processing methods on the other. It will soon be possible to investigate intact, three-dimensionally conserved cell nuclei on the level of individual nucleosomes. The ultimate goal would be to study the nanostructures of chromosomes in living cell nuclei changing in space and time. The new optical nanoscopic methods certainly have the potential to eventually carry out such research in four dimensions.
 HIGH RESOLUTION spectral precision distance microscopy reveals
three-dimensional positions of active and inactive ANT-genes on the female
X-chromosome.
HIGH RESOLUTION spectral precision distance microscopy reveals
three-dimensional positions of active and inactive ANT-genes on the female
X-chromosome.
((The active genes are found close to the surface, the inactive gene has moved into the interior and is invisible here))
((Source: Christoph Cremer, Peter Edelmann Heidelberg University and Sebastian Dietzel, Thomas Cremer Munich University))